To identify growth factor regulated genes, we have used gene traps in embryonic stem (ES) cells. In this approach, a promoterless reporter gene (for instance encoding βgalactosidase, or βgeo) is introduced in ES cells. Selection for expression of the gene requires transcription from a cellular promoter, and consequently a mutation in a cellular gene, and the activity of the tagged gene can be followed by staining for βgalactosidase activity. Detailed description of methods used for gene trap mutagenesis may be found in Friedrich and Soriano (1993), Chen and Soriano (2003), and Friedel and Soriano (2010). Large scale sequencing of ES cell clones was conducted and sequence tags have been deposited to NCBI’s dbGSS. A number of laboratories performing gene trap mutagenesis, including ours, have formed a gene trap consortium. Through the gene trap consortium, cell lines are made available to investigators at non-profit institutions, and may be ordered from the MMRRC. You can also search for our clones in the MMRRC catalog.
vector design
The basic gene trap vectors we have used include a reporter gene downstream of a splice acceptor sequence (Figure 1). They are therefore designed to function when inserted in an intron. The gene trap cassette is inserted in reverse orientation in a retroviral vector, and vectors and mutant lines derived from our screens are referred to as ROSA (Reverse-Orientation-Splice-Acceptor). Retroviruses insert as a single copy per locus, with no rearrangement of flanking sequences. They have a preference for insertions at the 5′ end of genes, often upstream of the initiator ATG, and the splice acceptor sequence we use does not appear to be bypassed. As a result, the majority of the mutations generated using our gene trap vectors are predicted to lead to null alleles. This has in fact been verified in all of the insertions we have analyzed to date at a molecular level
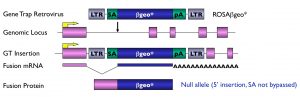 Figure 1: Gene Trap Design.
Figure 1: Gene Trap Design.
phenotypic analysis
Among 70 mutations that have been transmitted through the germ line (all called ROSA), about one third result in a recessive phenotype, either affecting embryonic development at different stages, or the adult.
| Line | Vector | Screen | Phenotype | Gene | Strain |
| SAβgeo 1-5 | SA βgeo | Random | 2/ 5 emb. lethal | Exoc4 (“Spock”) | SAβgeo 4 |
| ROSA 1-29 | ROSA βgeo | Random | 7/29 emb. lethal | Tead1 | ROSA 5 |
| 1/29 male sterile | Tpgs1 | ROSA 22 | |||
| Gt(ROSA)26Sor | ROSA 26 | ||||
| ROSA 30-39 | FUSA βgeo | Random | 7/10 emb. lethal | ||
| ROSA 40-45 | ROSA βgal | Induction (EB) | 1/ 7 emb. lethal | ||
| 1/ 7 male sterile | Bcl-W | ROSA 41 | |||
| ROSA 46-52 | ROSA βgal | Induction (TGFβ) | 1/ 7 emb. lethal | CD98 | ROSA 49 |
| ROSA 53-55 | ROSA βgalCre | Random | 1/ 3 emb. lethal | Shroom3 | ROSA 53 |
| ROSA 56-61 | ROSA βgeo* | Sequence | 1/ 3 growth ret. | HMG Box | ROSA 56 |
| 1/ 3 emb. lethal | Ctbp2 | ROSA 61 | |||
| ROSA 62-65 | ROSA βgeo* | Induction (RA) | 1/ 4 neuropathy | Sptbn4 | ROSA 62 |
| 1/ 4 male sterile | Map7 | ROSA 63 | |||
| ROSA 66-70 | ROSA βgal | Induction (serum) | 1/ 5 growth ret. | EFII | ROSA67 |
| AA09197 | ROSA68 | ||||
| ROSA 71 | ROSAFARY | Gene trap array | 1/13 emb. lethal | Strap | ROSA 71 |
| ROSA 72-83 | ROSAFARY | Gene trap array | 3/13 neonatal lethal | Zfand5, BC058969, Myo1e | ROSA72-83 |
| 9/13 postnat. phenotype | Arid5b, Zpf950, Schip1, Sgpl1, Tiparp, Csrnp1, Zfp640, Plekha1, Txnip |
induction trapping
To identify and mutate growth factor or retinoid regulated genes, we initially performed induction trapping by monitoring differential expression of the reporter gene, in ES cells or their differentiated derivatives. In some cases, such as ROSA49, we have replica plated ES cells infected with ROSA βgal and tested for differential lacZ activity upon exposure to exogenous factors. In other instances, such as ROSA62 or ROSA63, we have used a modified version of βgeo for the same purpose. βgeo encodes a βgalactosidase-neomycin phosphotransferase fusion protein and carries a mutation in the neo moiety that reduces its activity. This mutation has been corrected in βgeo*. As a result, induction trapping using ROSA βgeo* is more efficient as all neo resistant colonies are gene trap events yet only a fraction (~60%) exhibit βgalactosidase activity. We have also designed a novel vector system to isolate inducible gene traps by flow sorting (Medico et al., 2001).
It has been known for many years that activation of growth factor signaling pathways leads to the expression of multiple Immediate Early Genes (IEGs), yet their role in specifying particular growth factor responses remained controversial as similar IEGs are often engaged following induction of multiple signaling pathways. We have used gene trap-coupled microarray analysis, using a modified gene trap vector system in which 3′ RACE products from mutated genes are spotted on arrays (Chen et al., 2004), to identify targets of PDGF signaling, and their function in PDGF regulated processes. Mutations in these genes lead to a high frequency of phenotypes that affect the same cell types and processes as those controlled by PDGF signaling (Schmahl et al., 2007; Schmahl et al., 2008). These results suggest that these genes form a network that control specific processes downstream of PDGF signaling.
