
ROSAβgeo26 (ROSA26) refers to a locus that is widely used for achieving generalized expression in the mouse. The ROSAβgeo26 (GtROSA26) line was initially derived from pools of ES cells infected with the retroviral gene trap vector Gen–ROSAβgeo at low multiplicity of infection (Friedrich and Soriano, 1991). ES cells harboring a single proviral copy were injected into blastocysts and mice containing the ROSA26 insertion were isolated. Hence, ES cells representing the original infection event were never produced and ROSA26 ES cells have had to be generated de novo from transgenic animals. βgeo encodes a bifunctional lacZ/neomycin phosphotransferase gene and the ROSA26 strain is one of several strains that exhibits broad lac Z staining. However ROSA26 mice appear to express lacZ in all hematopoietic cells as well as in all tissue of the embryos (Zambrowicz et al,, 1997). It is thus very useful for a number of applications, including chimera analysis. ROSA26βgeo gene trap mice are viable as homozygotes but the recovery of these animals is very limited, particularly on a C57BL6/J background.
Please note that for larger embryos (E11.5 and above) and adult tissues, there is very inefficient penetration of the X-Gal stain. According to the tissue, penetration may be less than 1 mm. Please consult the X-Gal staining protocol for how to proceed in such circumstances.
The ROSA26 locus has been cloned and the provirus has been shown to interrupt two transcripts which encode a nuclear RNA (Zambrowicz et al., 1997). The characterization of this locus has led to two methods for generating transgenic mice that express a transgene is a generalized fashion. First, it is possible to use the genomic locus to target genes to the locus (Soriano, 1999). This has been used to produce a variety of knock -in lines, such a Cre reporter line expressing conditionally lacZ (GtROSA26tm1Sor; Figure 1; Soriano, 1999). A large number of knock-ins at this locus have been produced in many laboratories (for a partial, list, follow this link). These include reporter lines for site specific recombinases that activate a variety of different reporter genes, Flpe, Flpo, φC31o, estrogen regulated forms of Cre, DTA, luciferase and other genes (several of which are available from the Jackson labs). Our ROSA26Cre reporter mice (that express lacZ), available from the Jackson labs are readily recovered as fertile homozygotes, indicating that the difficulty in recovering GtROSA26 cannot be attributed to the mutation of the locus. For the second method, the promoter of the ROSA26 locus (pROSA26promoter) has been characterized in ES cells (Zambrowicz et al., 1997) and has been used for broad expression of transgenes in mice (Kisseberth et al., 1999). However in this case expression was broad but not uniform. Therefore in cases where uniform expression is desired, targeting to the locus might be the better way to go.
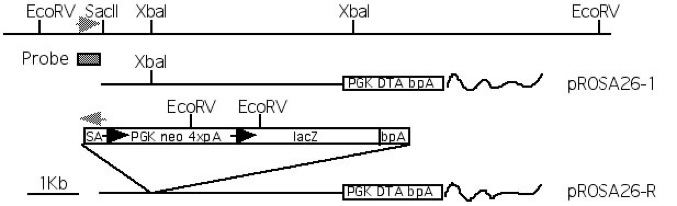
Figure 1. Restriction map and targeting of the Cre reporter construct to the ROSA26 locus. PCR primers from ROSA26 flanking (5’CCTAAAGAAGAGGCTGTGCTTTGG3′) and spice acceptor (5’CATCAAGGAAACCC TGGACTACTG3′) sequences were used to amplify a ~1.2Kb diagnostic fragment and are indicated by grey arrowheads. The probe used for Southern blot analysis is shown as a shaded box. LoxP sites are indicated by black arrowheads. Only EcoRV sites are indicated for pROSA26-R. For more details, see Soriano, 1999. pROSA26-1 sequence courtesy of Dr. Christophe Queva, AstraZeneca, Mölndal, Sweden.
Targeting to the ROSA26 locus (Figure 2) is conveniently achieved by introducing the desired gene into the first intron of the locus, at a unique XbaI site approximately 248 bp upstream of the original gene trap line (Figure 3). A construct using the same splice acceptor as used in the original gene trap allele (from adenovirus) followed by the gene of interest and polyadenylation site is inserted at the unique XbaI site of pROSA26-1. A neomycin resistance cassette is included in the targeting vector. ROSA26-1 already has a diphtheria toxin gene for negative selection and this allows high rates of homologous recombination (in our hands, ~25-50% of G418r clones). Targeted clones can be verified using a flanking probe (pROSA26-5′ or the 337 bp Not I fragment of pROSA26 promoter) and later genotyped using oligos derived from the locus flanking the insertion (Figure 3).
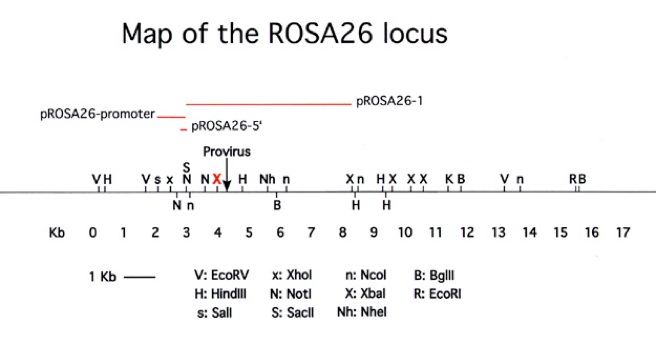
Figure 2. Diagram of the ROSA26 locus. The plasmids and the full sequence of pROSA26-5′, pROSA26-promoter and pROSA26-1 are available from the addgene repository (www.addgene.org). The insertion in pROSA26-5′ or the 337 bp Not I fragment of pROSA26-promoter can be used as probes to verify targeting events, but as the probes are short, washing should be performed at low stringency (1 x SSC at 65 degrees C).
5’=GGCTGTTTTGGAGGCAGGAAGCACTTGCTCTCCCAAAGTCGCTCTGAGTTGTTATCAGTAAGGGAGCTGCAGTGGAGTAGGCGGGGAG AAGGCCGCACCCTTCTCCGGAGGGGGGAGGGGAGTGTTGCAATACCTTTCTGGGAGTTCTCTGCTGCCTCCTGGCTTCTGAGGACCGCCC TGGGCCTGGGAGAATCCCTTGCCCCCTCTTCCCCTCGTGATCTGCAACTCCAGTCTTTCTAGAAGATGGGCGGGAGTCTTCTGGGCAGGC TTAAAGGCTAACCTGGTGTGTGGGCGTTGTCCTGCAGGGGAATTGAACAGGTGTAAAATTGGAGGGACAAGACTTCCCACAGATTTTCGGT TTTGTCGGGAAGTTTTTTAATAGGGGCAAATAGGAAAATGGAGGATAGGAGTCATCTGGGGTTTATGCAGCAAAACTACAGGTATATTGCTTGT ATCCGCCTCGGAGATTTCCATG(PROVIRUS)AGGAGATAAAGACATGTCACCCGAGTTTATACTCTCCTGCTTAGATCCTACTACAGTATGAAAT ACAGTGTYGCGAGGTAGACTATGTAAGCAGATTTAATCATTTTAAAGAGCCCAGTACTTCATATCCATTTCTCCCGCTCCTTCTGCAGCCTTATC AAAAGGTATTTAGAACACTCATTTTAGCCCCATTTTCATTTATTATACTGGCTTATCCAACCCCTAGACAGAGCATTGGCATTTTCCCTTTCCTGA TCTTAGAAGTCTGATGACTCA-3'
Figure 3. Sequence of the ROSA26 intron flanking the original provirus integration site. The XbaI site used for targeting genes to the locus is located 248 bp upstream and is indicated in bold and underlined. The 5′ oligonucleotides we use for PCR genotyping of the ROSA26βgeo gene trap strain and for the ROSA26 Cre reporter strain are indicated in red and blue, respectively. These oligos are used in conjunction with a reverse oligo from the ROSA26 sequence downstream of the provirus, indicated in green, and a reverse oligo from the SA sequence (5′-GCG AAG AGT TTG TCC TCA ACC-3′) using conditions described our PCR protocol.
